Endeavour Features: Molecular Structures
Endeavour Features Overview...
Previous: Space-group Determination...
Next: Simultaneous Optimization of Potential
Energy...
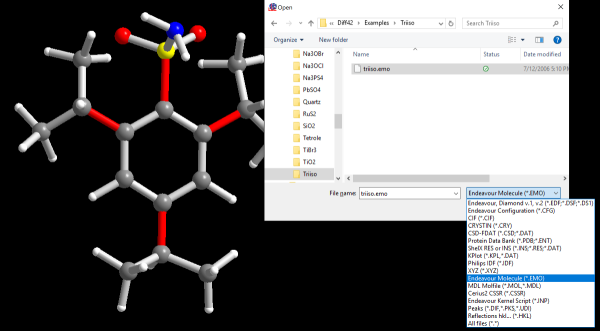
For molecular structures, it is generally extremely helpful to include any
knowledge of the molecular connectivity and/or 3D-structure of the molecule
itself in the structure solution process. These data can be imported from a
large variety of common molecular file formats (e.g. Cambridge CSD, Sybyl Mol2
etc.) as well as from known crystal structures.
During the structure solution process, the position and orientation of the
molecules as well as user-selected torsion angles within the molecule are
varied at random in order to find the correct structure solution.
You can fix molecules at certain positions and allow rotation along a reference
atom.
You can treat molecules as rigid-bodies (default setting in Endeavour 1.x) or as
flexible molecules (through the new advanced settings, available for users to change
since version 1.5). With rigid-body molecules, bond lengths within molecules cannot be varied during the structure solution
process.
Since version 1.8, atoms that belong to molecules may lie on special positions of the space
group (not possible in versions 1.0 - 1.7).
Endeavour Features Overview...
Previous: Space-group Determination...
Next: Simultaneous Optimization of Potential
Energy...
|

